Extracting quality DNA from marine invertebrates
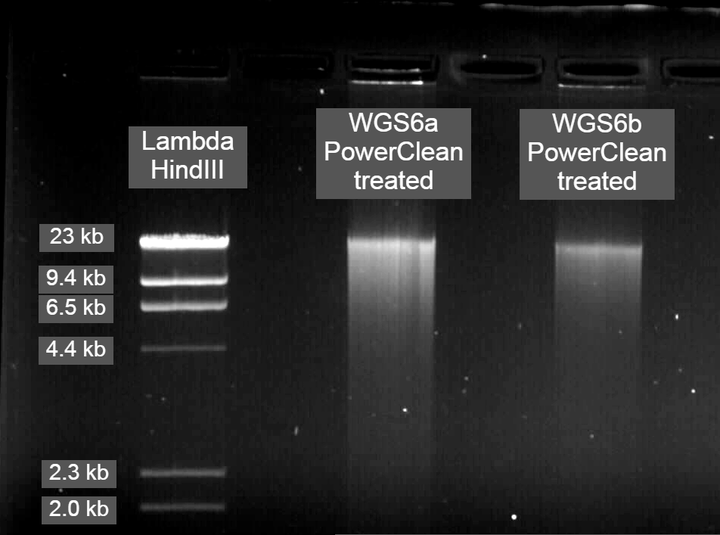 Genomic DNA submitted for PacBio sequencing
Genomic DNA submitted for PacBio sequencing
The problem
Extracting high molecular weight (HMW) DNA from marine invertebrates was one of the key challenges during my PhD. As I planned to use RAD sequencing to scan the genomes of the European lobster and the pink sea fan for variants, it was essential to obtain HMW DNA that was as pure and contaminant-free as possible.
I had samples of many different tissue types, including pleopods, walking legs, uropod v-notches and claw muscle from lobsters, and whole thumb-sized clippings from seafans. I quickly realised that this was not a trivial task as these tissue types often contain a lot of contaminants that are difficult to get rid of and interfere with the extraction protocol.
First attempts
At first I tried several kits available through various companies but these protocols typically produced poor results. So I went back to the basics and tried some phenol-chloroform, CTAB and salting-out protocols using freshly made reagents. After trying several protocols of each type, I still didn’t have much luck. Also, because I needed to extract DNA from hundreds to thousands of samples, I was not keen on the amount of extra time and effort required for handling phenol-chloroform!
The Salting-out protocol that keeps on giving
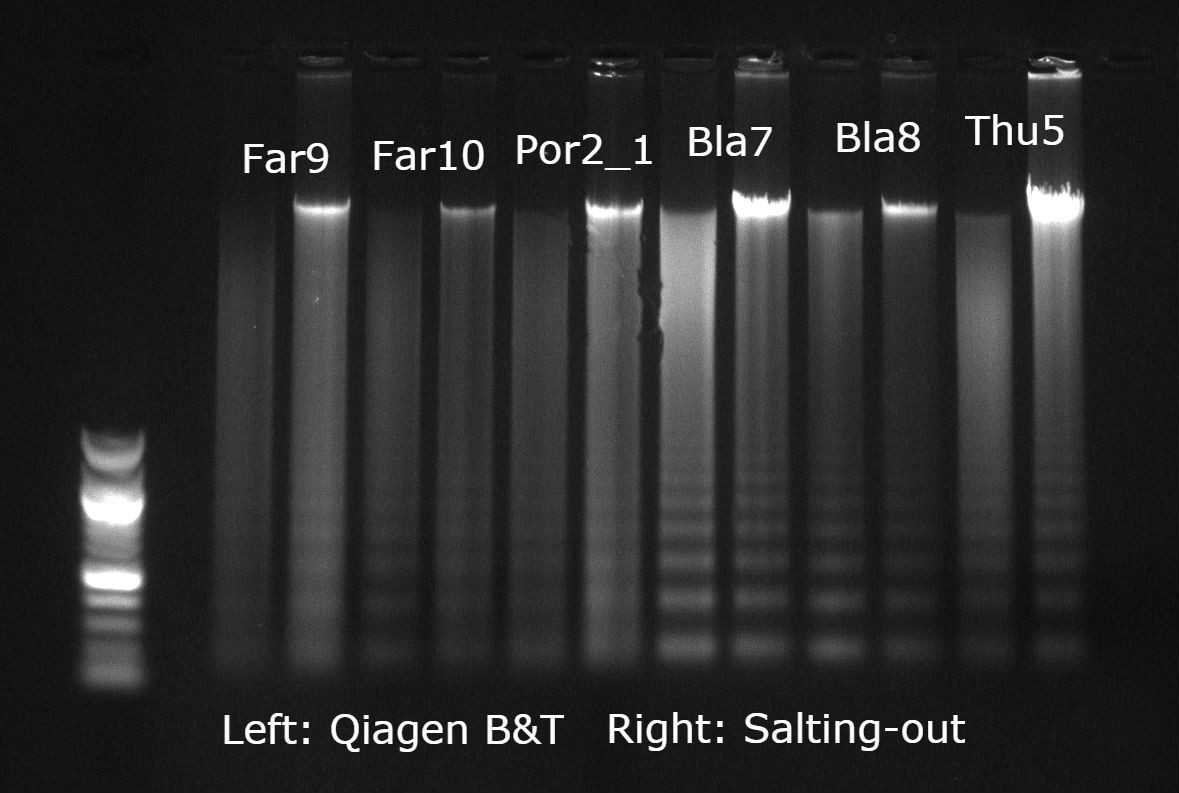
At this point, I thought all hope was lost until I came across a salting-out protocol that was specifically designed to extract DNA from crayfish exoskeletal tissue (i.e. pleopods and pereiopods). I modified this protocol to suit the lab equipment at my disposal and tested it on both the lobster tissues and the seafan tissue — the results were extremely encouraging! In fact, for the seafan, this was the first time I had managed to obtain DNA with a concentrated peak at around 20Kbp on a gel. The purity ratios on the Nanodrop were not bad either, typically at 1.75–1.85 (A260/280) and 1.90–2.20 (A260/230), although sometimes the latter ratio would drop < 1.5 but this tended to improve if I made the reagents up again fresh. I used this protocol to extract the majority of my samples and as I ended up with decent RAD sequencing data I think it is safe to say that overall it worked rather well! A few of my colleagues tried out this method on other marine invertebrates, including mussels and other soft corals, and also got similar results. Check out the protocol at the end of this post.
Gel image example
PowerClean CleanUp protocol improved purity
For PacBio library preparation and sequencing, I was advised by one of the reps to clean the DNA post-extraction using the DNeasy PowerClean CleanUp Kit. For obtaining very pure DNA that was contaminate-free, this proved to work extremely well and produced much more consistent results on the Nanodrop than the salting-out protocol alone. Therefore, my advice to others is if you are processing many samples then stick to the salting-out protocol as this should work well if you are using RAD sequencing or another reduced representation sequencing technique. However, if you have a relatively small number of samples or are doing long-read PacBio sequencing, I recommend the workflow below.
Recommended workflow for PacBio sequencing
Download protocols
Salting-out protocol
DNeasy PowerClean CleanUp Kit
Twitter thread
In my experience, extracting quality DNA from marine invertebrates is extremely challenging.
— Tom Jenkins (@Tom__Jenkins) August 17, 2020
Therefore, I've written a short post on what I did to help overcome these challenges – hope it's useful!#AcademicTwitter #popgen #phdchat https://t.co/om9Nl2t376